Lupine Publishers | Journal of Biotechnology
Abstract
Antimicrobial resistance (AMR) is a growing problem worldwide.
Resistance to antibiotics can occur in a number of ways, one
of which is removal of the drugs from the cell via efflux pump
macromolecular machineries. As such, efflux pumps can provide a
background level of resistance to many different classes of
antimicrobials and are a major contributor to AMR. Inhibition of efflux
pumps therefore has the potential to reverse resistance to many
antibiotics in one go and is an attractive potential for treating
resistant infections. Whilst a number of efflux inhibitors are known,
none are currently used clinically due to harmful side effects.
Development of novel inhibitors is therefore imperative. The article
aims to review accumulation assays and efflux assays, two of
the most common laboratory techniques used to identify and characterise
candidate efflux inhibitors.
Keywords:Efflux pumps; Efflux inhibitors; Efflux assays; Antimicrobial resistance; Drug discovery
Introduction
Globally, antimicrobial resistance is a rising public health
challenge. Particular infections including pneumonia, Tuberculosis
(TB), gonorrhoea, and salmonellosis are becoming more difficult
to treat. Of new TB cases, 3.5% are either resistant to rifampicin
(the most effective first line drug) or are multi-drug resistant,
rising to 18% for previously treated individuals [1] Furthermore,
there are fears that Neisseria gonorrhoeae has already developed
resistance to all currently recommended treatments [2]. There is
a desperate need for new antibiotics to treat these most resistant
of infections, but the huge costs, long timescale and high attrition
rate of drug discovery means that this is a slow process. Twenty
classes of antibiotics were discovered between 1940 and 1962,
yet only two have been developed since then [3]. Moreover, for
any novel antibiotic developed, it is likely that resistance will
quickly emerge once it is brought into clinical use, especially
with the frequent misuse of antibiotics which drives selection for
resistance. Therefore, other strategies must be taken in parallel to
antibiotic development, or there will be a continuous arms race of
drug development and resulting gain of resistance, a battle we are
currently losing.
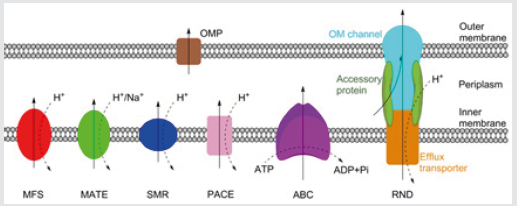
Figure 1: Schematic representation of the MFS, MATE, SMR,
PACE, ABC and RND families of bacterial efflux pumps, plus
an outer membrane protein channel (OPM), shown here in a Gram-negative
bacterium. RND family efflux pumps comprise
of a tripartite complex formed from an inner membrane efflux
transporter, an outer membrane channel, and a periplasmic
accessory protein. All six families, with the possible exception of the
PACE family, also have representatives in both Grampositive
and acid-fast bacteria. Bold arrows indicate the direction of drug
efflux, and dashed arrows show ion movement.
Antibiotic resistance can occur via acquired or intrinsic
mechanisms. Acquired resistance, typically via horizontal transfer
or spontaneous mutation, often functions by altering the drug
target or production of enzymes which degrade the antibiotic.
Acquired resistance, gained in response to antibiotic treatment, is
usually only effective against a single drug. Intrinsic resistance, on
the other hand, refers to the non-specific mechanisms of antibiotic
resistance evolved ancestrally, including the impermeable outer
membrane of Gram-negative or acid-fast group of bacteria, and
drug efflux pumps which remove drugs from the cell [4]. There are
currently six families of bacterial efflux pumps identified: the ATPBinding
Cassette (ABC) family, the Major Facilitator Superfamily
(MFS), the Multidrug And Toxin Extrusion (MATE) family, the Small
Multidrug Resistance (SMR) family, the Resistance-Nodulation-Cell
Division (RND) superfamily and the Proteobacterial Antimicrobial
Compound Efflux (PACE) family, which has not yet been structurally
characterised. The ABC family hydrolyse ATP directly to drive
efflux, whereas the other five utilise transmembrane ion gradients
[5]. Whereas the RND family directly effluxes antibiotics across
both membranes, the other five families only transport antibiotics
across the inner membrane. From the periplasm, drugs can exit the
cell via outer membrane protein channels or by entering the RND
complex (Figure 1).
Efflux pumps are often non-specific, and as such can provide
resistance to a wide range of antimicrobials. They have been
implicated in contributing towards the multi-drug resistant
phenotypes of Mycobacterium tuberculosis [6], Pseudomonas
aeruginosa [7], Neisseria gonorrhoeae [8], and Streptococcus
pneumoniae [9], amongst others. Inhibition of drug efflux is
therefore an exciting prospect for treating drug resistant bacteria
and may enable old antibiotics to re-enter clinical usage. There is
compelling evidence that the use of efflux pump inhibitors as an
adjuvant may aid treatment of resistant infections of many types
[6-12]. However, despite a number of potent efflux inhibitors
being known, none have entered clinical use. In most cases this is
because the compounds are toxic at the concentrations required
to inhibit efflux [13]. There is therefore a pressing need to develop
novel clinical efflux inhibitors. To achieve this, assays are needed
to validate the inhibitory activity of novel compounds. One way
this can be achieved is by using standard antibiotic susceptibility
testing, such as the resazurin-based microplate assay to determine
if the putative inhibitor, at sub-MIC concentrations, is able to lower
the MIC of a known antibiotic. This method has the benefit of being
relatively easy and high-throughput; furthermore, it is possible to
combine this method with mutants of efflux pumps to confirm that
the effect on the MIC is occurring specifically via inhibiting efflux,
and even to identify which efflux pump is inhibited [14]. However,
using reduction of MICs to identify and validate efflux inhibitors
is fairly insensitive, and so is of limited use. Only large changes
to efflux will likely have an effect on MICs, and so less potent
inhibitors may be dropped out. Furthermore, as this method does
not measure efflux, it is difficult to directly attribute changes in MIC
to efflux inhibition [15]
A more direct way is therefore needed to study the effect of
candidate inhibitors on efflux. One way is to follow the movement
of an efflux pump substrate, often a fluorescent molecule, into and
out of bacterial cells, and use this as a measure of efflux activity.
Many different molecules are used to measure efflux, with ethidium
bromide and Nile red being two of the most common. Ethidium
bromide fluoresces strongly when bound to DNA, and Nile red
fluoresces when in non-polar environments such as the membrane
[16,17]. This therefore gives these molecules the advantage that they
fluoresce differentially in extra- and intracellular environments,
providing a sensitive indication of rate of efflux from the cell, and
helping eliminate background fluorescence. These methods fall
into two main categories; those which follow the accumulation of
the molecule within the cell, and those which follow its efflux.
Accumulation Assays
Whilst there are variations, most accumulation assays typically
follow a similar procedure. At the start of the assay, there is no
dye added to the bacteria. This is then added to the reaction, and
its accumulation within the cells followed over time, typically by
measuring the fluorescence with dyes such as ethidium bromide.
Eventually, accumulation will tail off, with fluorescence reaching a
steady state. This reflects an equilibrium being achieved between
influx and efflux of the dye. This assay can be performed with added
efflux inhibitors [18]. By inhibiting efflux, more dye accumulates
within the cells compared to untreated ones, with steady state
being achieved at a higher fluorescence. This assay can therefore
be used as a very simple test to validate the inhibitory activity of
a candidate efflux inhibitor [19]. Similarly, accumulation assays
are often used to observe changes in efflux ability in knockout,
knockdown or overexpression mutants.
If a knockout/knockdown mutant accumulates more dye, it can
be assumed that the gene encoded a protein important for drug
efflux, or a regulator of these, and vice versa with overexpression
mutants. These two approaches can be combined, with different
mutants treated with efflux inhibitors to see if they have a greater
or lesser effect on dye accumulation than for wild-type cells. This
can help determine which efflux pump the inhibitor affects [7].
However, there are problems with using accumulation assays, the
most important being that accumulation is not a direct measure
of efflux. Rather, it reflects a number of factors, predominantly
the balance of influx and efflux rates. Influx depends greatly on
the permeability of bacterial membranes, which can vary greatly
between even closely related strains due to differing membrane
compositions [20]. Therefore, unless influx rates are known,
kinetic data cannot be obtained from accumulation assays and
results remain qualitative. Whilst this limits usage of accumulation
assays to comparisons between isogenic mutants, or groups treated
with different inhibitors, the assay remains a conclusive way to
determine if a molecule possesses inhibitory activity, and so is
frequently used to validate new efflux inhibitors.
Efflux Assays
If a quantitative measure of efflux is required, then a more
direct efflux assay should be used. This follows a similar premise to
accumulation assays, but instead involves preloading the cells with
dye and following its subsequent efflux. To achieve this, cells are
incubated with a dye or other efflux pump substrate, and a known
efflux inhibitor such as CCCP. This causes the dye to accumulate to a
maximum level. Then, the cells are washed to remove the inhibitor
and any remaining extracellular dye. The cells are then reenergised,
typically with glucose, which restarts efflux. The movement of the
dye out of the cells can be followed by recording the decreasing
fluorescence [15]. As this method is a direct measure of efflux, kinetic
data can be obtained for efflux rates, which allows comparisons to
be made more broadly, rather than just between isogenic species.
In much the same way as with accumulation assays, modifications
can be made to study the effects of putative inhibitors or different
mutations on efflux rates [12,21].
Efflux assays are very sensitive, and they allow for validation
and characterisation of novel inhibitors, which may potentially
have clinical usage. Whilst the efflux assay is widely used, it is not
always applicable. Non-fermenter bacteria, including Pseudomonas
and Acinetobacter, are unable to metabolise glucose, and so
cannot be easily reenergised. This means that efflux assays can be
unsuitable for some bacteria, and instead accumulation assays are
more commonly used [7,22].
Limitations with these Assays
A fundamental problem with both types of assay is that using
ethidium bromide or another dye to measure efflux or accumulation
is of limited clinical relevance, and may not reflect well the efflux of
any particular antibiotic. This can be due to the dye and antibiotic
having very different kinetics of efflux, and furthermore, they may
not even be substrates for the same efflux pumps. In addition, as
ethidium bromide intercalates with DNA, there is a lag time in efflux
in which it dissociates, followed potentially by a two-step efflux
mechanism in which it is first transported to the periplasm. This can
lead to underestimates of efflux rate, and so may be a poor reflection
of efflux rates of antibiotics [23]. Therefore, where possible, it is
better to use the antibiotic of interest itself as a direct measure of
efflux, although this tends to be far more difficult experimentally.
Certain antibiotics, such as fluroquinolones and tetracyclines have
endogenous fluorescence which enables their accumulation to be
followed [24]. For non-fluorescent antibiotics, Mass-Spectroscopy
(MS) can be used to directly study their accumulation. A recent
proposed joint protocol for spectrofluorimetric and MS analyses
suggests that the two methods are complementary and together can
accurately measure antibiotic accumulation, demonstrated with
fluroquinolones [25]. MS analyses, rather than spectrofluorometric,
may also provide a better way to screen natural compounds
for efflux inhibitory activity. Many natural compounds have
endogenous fluorescence, which can make it hard to isolate and
interpret fluorescence changes due to dye accumulation or efflux.
As before, the actual antibiotic, rather than a dye, could be used, and
MS used to determine how much accumulates with and without the
candidate inhibitor.
One of the biggest problems facing the development of novel
efflux inhibitors is the lack of high-throughput assays to validate
putative compounds. Whilst both the accumulation and efflux assays
are relatively easy to perform and can reliably confirm if inhibition
occurs, both are limited on throughput. Therefore, whilst some in
silico screening has been performed [26], limitations in throughput
have so far prevented large-scale screening of libraries in vitro.
Instead, the search for novel inhibitors has relied extensively on
prior knowledge to select candidates for validation. Whilst the hit
rate with this has been relatively high, the overall number of new
inhibitors found has been low, and it is rare to identify completely
novel inhibitors in this way. This is in part why no inhibitors have
made their way into clinical usage, as many are closely related and as
such are similarly toxic. Development of high-throughput screening
assays for novel inhibitors is therefore necessary if efflux inhibitors
are to progress clinically. Recently, the Back assay was developed,
which uses a 96-well plate format combined with MS. This was able
to test in triplicate 12 compounds at 4 concentrations each, for two
different Escherichia coli strains [27]. This progression to more
high-throughput screening is likely to be the driving force behind
development of novel efflux inhibitors, and further work needs to be
done to optimise assays before large scale-screening of compound
libraries can be performed. Ultimately, the development of clinical
efflux inhibitors used therapeutically as antibiotic adjuvants may
be what turns the tide in the battle against antibiotic resistance.
Acknowledgment
The authors would like to thank the British Society for
Antimicrobial Chemotherapy, without whose funding this work
would not have been possible. We also wish to thank Dr Arundhati
Maitra for her time and advice when writing the article, as well as
for help with ChemBioDraw
Read more Lupine Publishers Blogger Articles please click on: https://lupinepublishers.blogspot.com/
Read more Biotechnology & Microbiology Journal blogger articles please click on: https://lupine-biotechnology-microbiology.blogspot.com/

No comments:
Post a Comment